Research
Growth and reproduction of organisms are cell cycle dependent. The cell cycle consists of two main parts: the interphase, in which the DNA is replicated, followed by the mitosis phase, where the duplicated sister chromatids are segregated to the newly formed daughter cells. The mitotic phase itself is divided into 5 distinct phases, finally leading to the faithful separation of the daughter cells, which is also known as cytokinesis (see Fig. 1). While the interphase can take up to 16-20 hours, mitosis and cytokinesis finish in a relatively short period of time (usually within 1 hour), making it more challenging to monitor the underlying mechanisms. The main interest of our lab is to illuminate regulatory events during mitosis and cytokinesis, which are also termed as cell division.
The underlying dynamics of structural and biochemical changes during cell division are still poorly understood. The major questions that we aim to answer are not only related to alterations within dividing cells, but also to the reshaping processes of the cell surface in coordination with the cell’s interior. Also, we aim to elucidate how the cell is communicating with its extracellular environment during division. Another important aspect for us is the role of post-translational modifications, in particular phosphorylation events, which are known to drive cell cycle progression.
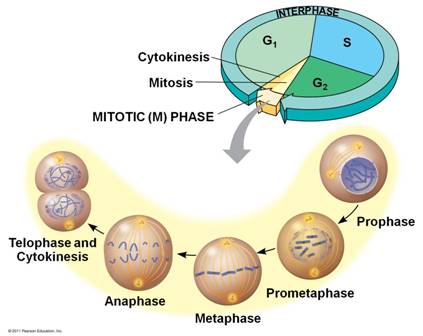
Fig 1. Cell cycle phases (www.quio.com).
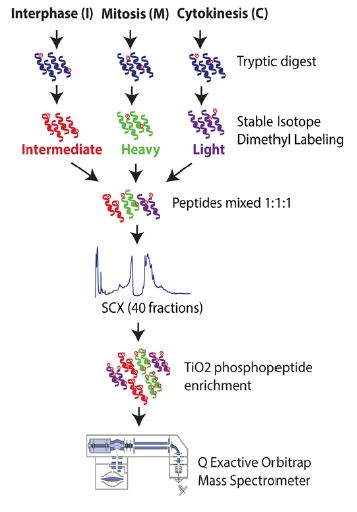
Fig 2. Proteome analysis of cell cycle stages (Karayel et al., 2018. Sci Rep 8: 2269-85).
To examine the biochemical profile of different cellular components as mammalian cells progress through mitosis and cytokinesis, we apply microscopy based imaging techniques. In particular, we investigate the functional changes of regulatory factors under physiological conditions and in response to inhibitory drugs. We are also interested in the proteome and phosphoproteome profiles of dividing or migrating cells. For this purpose, we apply quantitative proteomic techniques such as SILAC or dimethyl labeling followed by LC-MS/MS analysis (see Fig. 2). Moreover, finding interaction partners of regulatory proteins of the cell division machinery is one of our main goals. Herefore, our lab has expertise in BioID experiments followed by mass spectrometry based analysis. Furthermore, we developed bioinformatics pipelines to statistically and computationally analyze proteome and phosphoproteome data. Our focus lies on the identification of conserved motifs of phosphosites and the generation of kinase-substrate networks of phosphorylation events.
